Article published in Naure details, that by using medical text sources entered into electronic medical records, it is possible to learn about possible diseases such as viral pneumonia.
The article "Real-time clinician text feeds from electronic health records", 'published in Nature in February this year, explains how search engine and social network analysis have been attempted for outbreak detection of infectious diseases.
"We describe an approach using real-time aggregation of keywords and phrases of freetext from real-time clinician-generated documentation in electronic health records to produce a customisable real-time viral pneumonia signal providing up to 4 days warning for secondary care capacity planning," the authors explain. This is a low-cost, open-source approach that is ideal for local customization as it does not require any specific clinical record system.
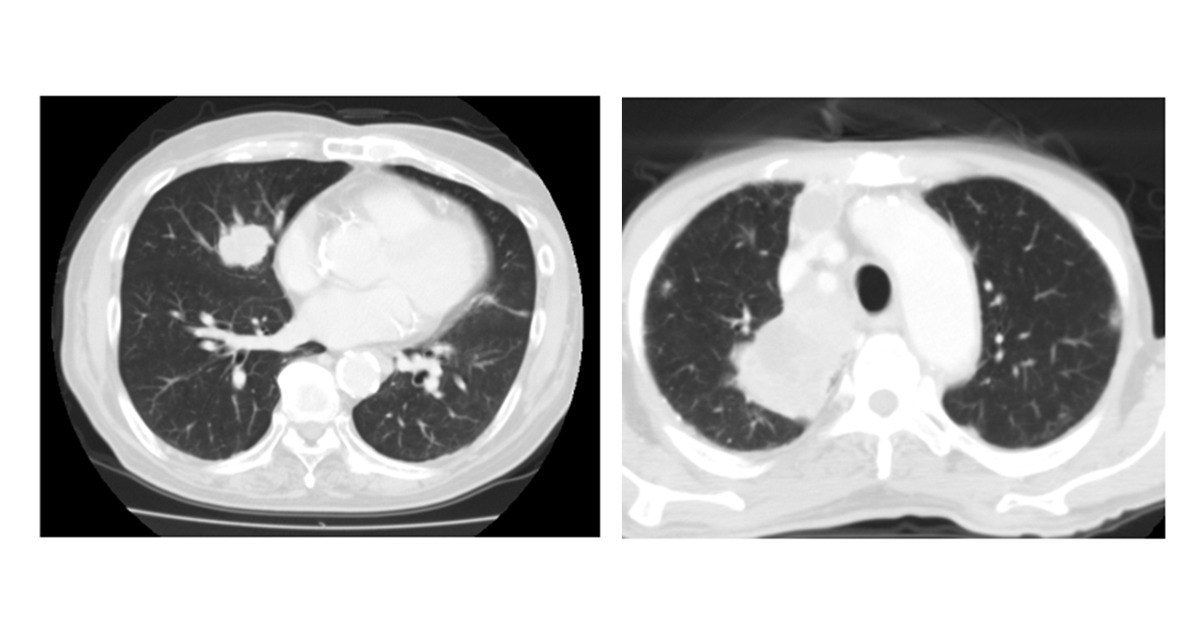
The same approach of real-time aggregation of keywords and phrases has been used to detect possible cases of COVID, classifying the symptoms of symptomatic cases such as: "dry cough", "pyrexia", "fever", "dyspnea", "anosmia", "pneumonia", among others.

The system was applied at King's College Hospital and Guys & St Thomas Hospital in the United Kingdom. In both hospitals, an increase in COVID-19 positive PCR tests was detected almost simultaneously with COVID-19 symptom-related keywords added to the hospital's patient records.
However, despite the important findings the scientists explain that this approach has limitations, such as the environment in which it was applied, or the handling of the data. "this study as proof-of-concept was confined to secondary care settings; during the Covid first wave in March 2020, there were triage mechanisms to redirect low severity cases away from secondary care affecting visibility. For a global view of a health economy, text aggregators would also need to be deployed in primary care and emergency care providers."
Read the full text at the following link: https://www.nature.com/articles/s41746-021-00406-7